Beyond the helix: new insights into the structure, function and design of RNA therapeutics using biophysics technologies and deep learning
EPSRC CASE Studentship (collaborative landscape award)
Deadline: 29th January 2025. Fully funded position with stipend, open to home students only.
About the Project
Therapeutic RNAs and vaccines are being developed for a broad range of human diseases. However, their optimization is hindered by mRNA instability and inefficient protein expression. mRNA secondary structure plays an integral role in the biosynthesis of proteins [1-2] as well as molecule stability [3]. Therefore, the higher order structure of RNA has an important role on the biological activity. There is currently a significant lack of robust analytical methods or biophysical approaches that provide significant insight into mRNA structures.
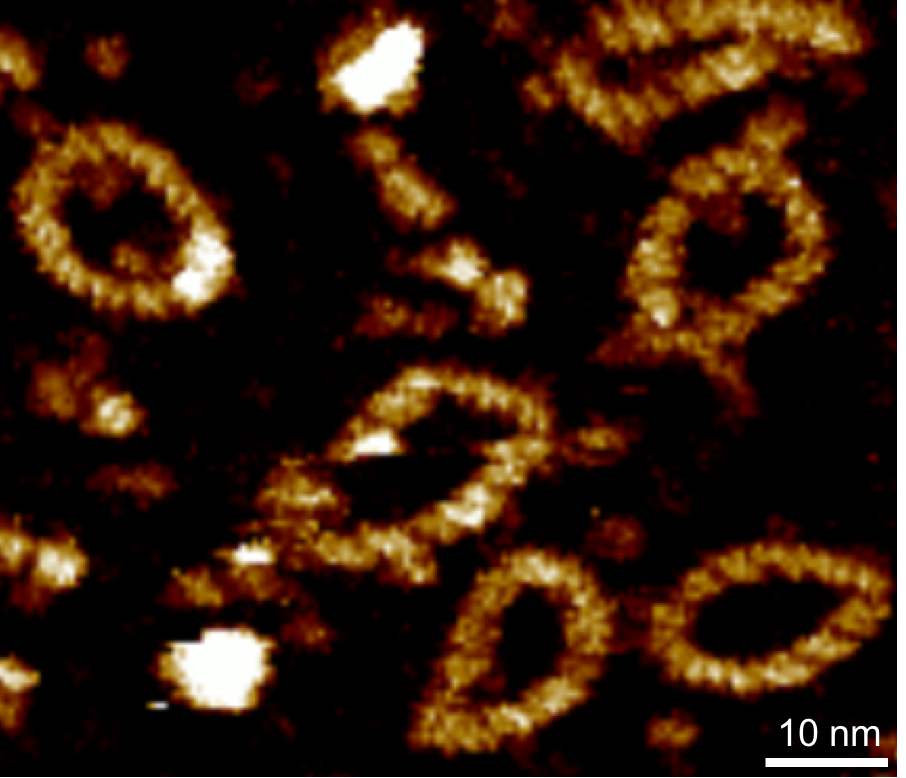
We have developed high-resolution atomic force microscopy (AFM) methods that are unique in their ability to provide quantitative information on nucleic acid structure, function, and interactions in liquid with sub-molecular resolution without the need for labelling or averaging [4-5]. Our preliminary data shows that we can achieve equivalent resolution on RNA molecules, which forms the basis of this ambitious project in collaboration with AstraZeneca.
We have developed a single-molecule analytical pipeline which combines atomic force microscopy (AFM) and deep-learning image analysis methodologies to quantify the formation of complex nucleic structures with nanometre resolution [5-6]. Our pipeline can discriminate between molecular topologies (e.g. linear vs circular), and determine molecular shape, conformation, and aggregation for individual DNA molecules within a larger population.
This proposal will be a collaboration with AstraZeneca to develop a novel pipeline for the structural and functional characterisation of novel mRNA therapeutics using high-resolution, single particle, atomic force microscopy (AFM) imaging in combination with optical tweezers and HPLC [7]. Our pipeline has the potential to accelerate research and development into RNA therapeutics, providing currently inaccessible structural data showing changes in the secondary and tertiary structure of RNAs. We can carry out these measurements at distinct time points after assembly, providing insight into the stability of RNA structures. We will obtain automated quantitative measurements of structure, aggregation and morphology, and link these back to changes in manufacturing of formulation design, allowing AZ to link quantitative structural parameters to their manufacturing processes in a new, faster feedback loop.
This project will aim to answer the fundamental question, how does mRNA structure impact protein production?
The objectives are to:
• Use biophysical methodologies to determine the structure and stability of a test set of mRNA molecules, which have different predicted structures but encode for the same protein
• Develop a pipeline to build up our understanding of structure function relationships in mRNA by bridging biophysical structural data to functional data from other methodologies
• Test this pipeline on new emerging mRNA candidates to determine whether we can improve the mRNA manufacturing feedback loop
Candidate Requirements
Candidates must have achieved a minimum 2:1 undergraduate and/or postgraduate masters’ qualification (MSc) in Chemical Engineering, or a relevant subject, by the start of the PhD. The English language requirements (overall IELTS grade of 6.5 with a minimum of 6.0 in each component, or equivalent) must also be met by the start of the PhD.
How to Apply
Please refer to the EPSRC DLA webpage for detailed information about the EPSRC DLA and how to apply.
Funding Notes
The award will fund the full (UK) tuition fee and UKRI stipend (currently £19,237 per annum) for 4 years, as well as a research grant of £4500 to support costs associated with the project.
References
1.Gaspar P et al., NAR 2013 https://academic.oup.com/nar/article/41/6/e73/2902446
2.Mauger DM et al., PNAS 2019 https://www.pnas.org/doi/abs/10.1073/pnas.1908052116
3.Wayment-Steele HK et al., NAR 2021 https://academic.oup.com/nar/article/49/18/10604/6370252
4. Pyne ALB. et al., Nat. Commun. 2021 https://www.nature.com/articles/s41467-021-21243-y
5. Holmes, EP et al., BioRxiv 2024 https://www.biorxiv.org/content/10.1101/2024.06.28.601212v2
6. Beton JG et al., Methods, 2021. https://pubmed.ncbi.nlm.nih.gov/33548405/
7. Nwokeoji AO. et al., Analyst, 2019. https://pubs.rsc.org/en/content/articlelanding/2019/an/c9an90100k