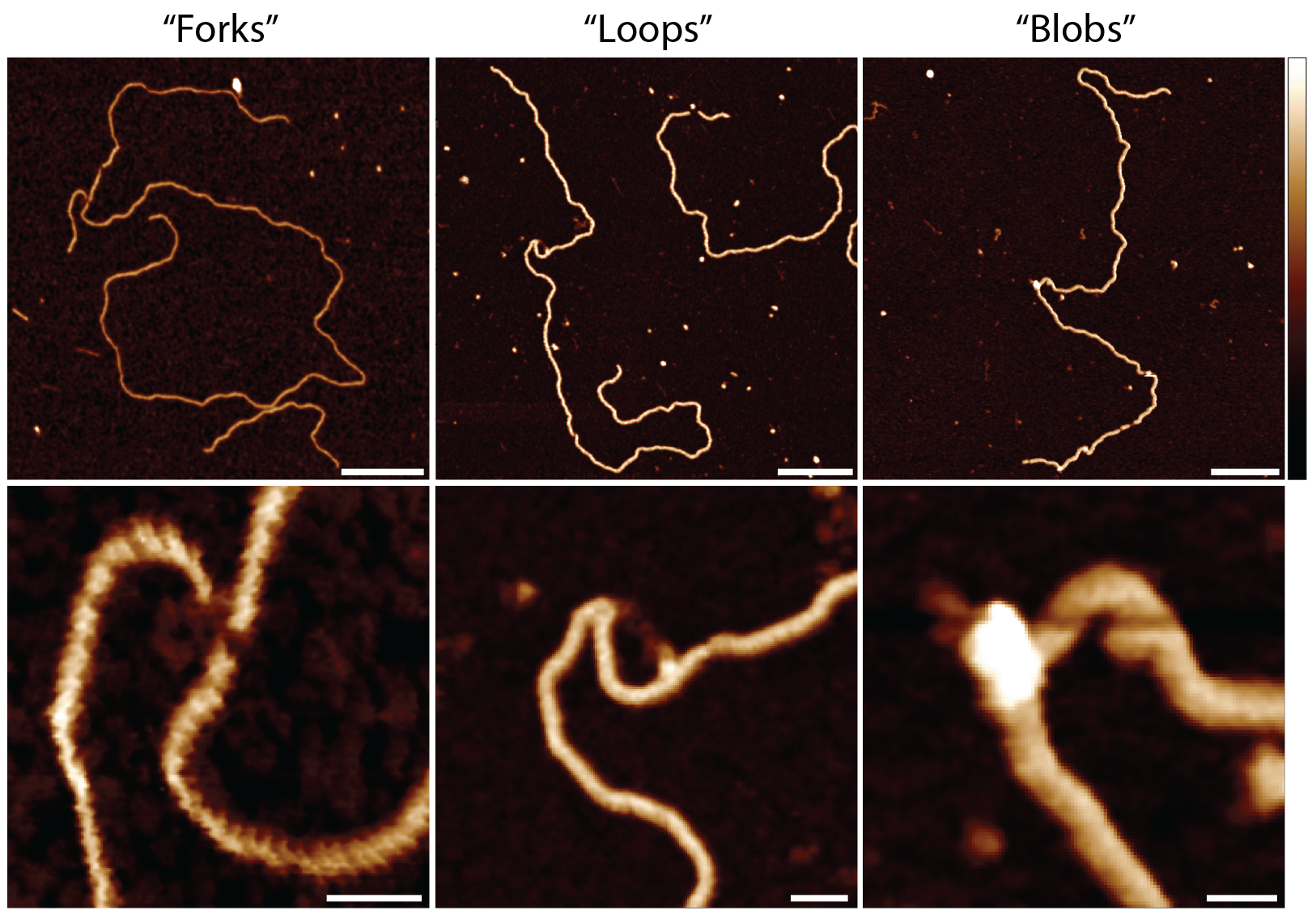
Our latest research on the “Oligomerisation of Ku from Mycobacterium Tuberculosis promotes DNA synapsis” has recently been published in Nature Communications. In this work we explored the importance of the bacterial protein, Ku, on forming DNA-Protein oligomers. This DNA-protein oligomerisations points towards the mechanisms of the vital DNA repair mechanism in Mycobacterium Tuberculosis, which ultimately is responsible for the spread and rising cases of Tuberculosis. This research poses an exciting insight into the causes of rising issues with antimicrobial drug resistance and highlights a potential target for therapeutics. This work was done as part of a collaboration with Dr Amanda Chaplin and Dr Sayma Zahid from the University of Leicester.
The full paper can be found here.